Autophagy-regulated Signalosomes in Cellular Stress and Disease Pathways (July 2016-July 2021)
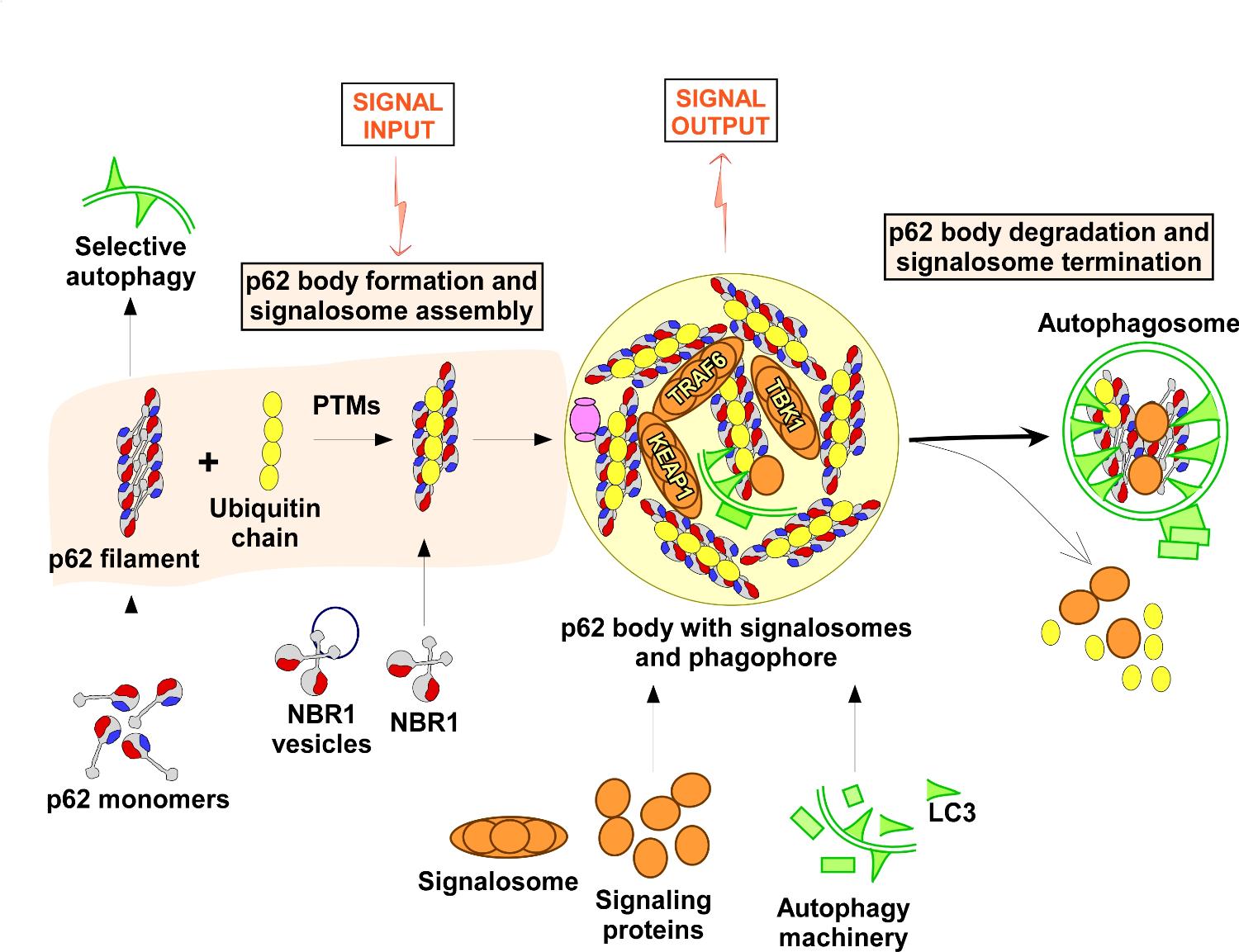 Illustration of the dynamics of p62 mediated signalosome formation and degradation. p62 filaments are continuosly degraded by selective autophagy. In response to a stress signal, post-translational modifications (PTMs) induces interactions between p62 filaments and ubiquitin chains resulting in the transient formation of p62 bodies and a p62 dependent assembly of specific signalosomes. After an active signaling phase, a coordinated termination of the signalosomes is secured by deubiquitination (DUB)-mediated disassembly of p62 bodies and selective autophagy of p62 along with selected signaling components.
Illustration of the dynamics of p62 mediated signalosome formation and degradation. p62 filaments are continuosly degraded by selective autophagy. In response to a stress signal, post-translational modifications (PTMs) induces interactions between p62 filaments and ubiquitin chains resulting in the transient formation of p62 bodies and a p62 dependent assembly of specific signalosomes. After an active signaling phase, a coordinated termination of the signalosomes is secured by deubiquitination (DUB)-mediated disassembly of p62 bodies and selective autophagy of p62 along with selected signaling components.
Vision and Aim
Our main hypothesis is that the selective autophagy receptor p62/SQSTM1 regulates cellular stress signalling through both formation and degradation of stress-induced cellular bodies called signalosomes.
Our vision is to profoundly increase the current knowledge on selective autophagy receptors acting both as signaling hubs and signaling terminators, thereby integrating and regulating cellular stress signal pathways. The knowledge generated will help identify novel therapeutic targets for neurodegenerative diseases, cancer and inflammation. The new findings will also be highly relevant for increased understanding of innate and adaptive immunity, infectious diseases involving intracellular pathogens and aging.
Our work on the scaffold and signaling protein p62/SQSTM1 (sequestosome-1) led to the discovery of selective autophagy at the molecular level (1,2). Our two first papers describing p62/SQSTM1 as a selective autophagy substrate and receptor for degradation of ubiqutinated substrates are each cited more than 1000 times. With this 5-year basic research program we aim to firmly establish our research group at the forefront of international research on selective autophagy, and in particular research on the role of selective autophagy in regulation of cellular signaling.
Toppforsk Fellesløftet III
56 published papers 2017-2021
•11 original research papers from our group
•20 review papers (minireview and large reviews)
•3 methods papers
•22 papers with coauthorship on papers from groups we have collaborated with
•Total of 56 papers in the 5-year project period
•Journals: Autophagy, Annual Reviews in Cell and Developmental Biology, EMBO Journal, EMBO Reports, Journal of Cell Biology, Nature Communications, Nature Cell Biology, Nature, Molecular Cell, Developmental Cell, Cell Reports, Journal of Cell Science, Journal of Molecular Biology